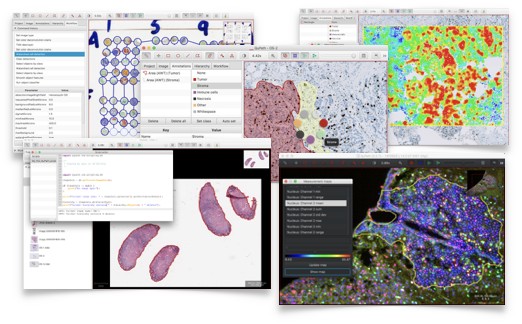
QuPath is open source software for bioimage analysis.
Features include:
- Lots of tools to annotate and view images, including whole slide & microscopy images
- Workflows for brightfield & fluorescence image analysis
- New algorithms for common tasks, including cell segmentation, tissue microarray dearraying
- Interactive machine learning for object & pixel classification
- Customization, batch-processing & data interrogation by scripting
- Easy integration with other tools, including ImageJ
To download QuPath, go to the Latest Releases page.
For documentation, see https://qupath.readthedocs.io
For help & support, try image.sc or the links here
To build QuPath from source see here.
If you find QuPath useful in work that you publish, please cite the publication!
QuPath is an academic project intended for research use only. The software has been made freely available under the terms of the GPLv3 in the hope it is useful for this purpose, and to make analysis methods open and transparent.
QuPath is being actively developed at the University of Edinburgh by:
Past QuPath dev team members:
- Melvin Gelbard
- Mahdi Lamb
For all contributors, see here.
This work is made possible in part thanks to funding from:
- Wellcome Trust Technology Development Grant (2022-Present)
- CZI Essential Open Source Software for Science, Cycle 4 (2022-Present)
- CZI Essential Open Source Software for Science, Cycle 1 (2020-2022)
- Wellcome Trust / University of Edinburgh Institutional Strategic Support Fund (ISSF3) (2019-2020)
QuPath was first designed, implemented and documented by Pete Bankhead while at Queen's University Belfast, with additional code and testing by Jose Fernandez.
Versions up to v0.1.2 are copyright 2014-2016 The Queen's University of Belfast, Northern Ireland. These were written as part of projects that received funding from:
- Invest Northern Ireland (RDO0712612)
- Cancer Research UK Accelerator (C11512/A20256)