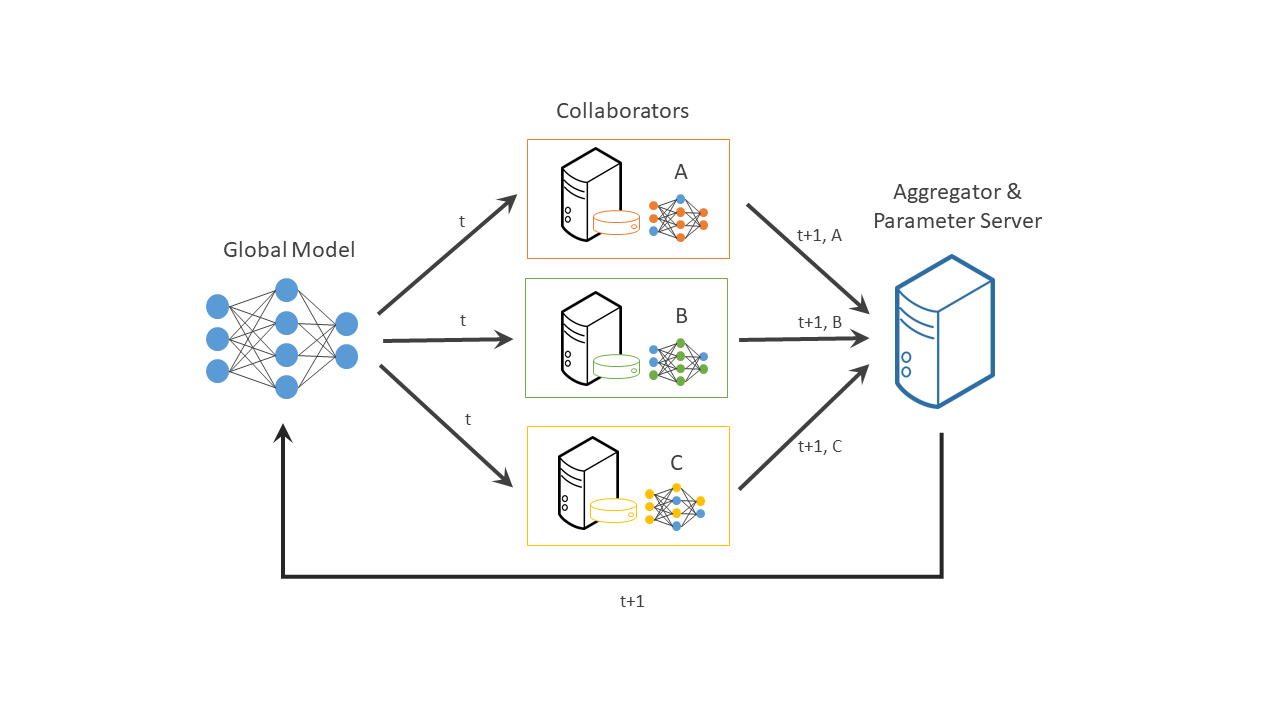
OpenFL is a Python 3 framework for Federated Learning. OpenFL is designed to be a flexible, extensible and easily learnable tool for data scientists. OpenFL is developed by Intel Internet of Things Group (IOTG) and Intel Labs.
You can simply install OpenFL from PyPI:
$ pip install openfl
For more installation options check out the online documentation.
OpenFL enables data scientists to set up a federated learning experiment following one of the workflows:
-
Director-based Workflow: Setup long-lived components to run many experiments in series. Recommended for FL research when many changes to model, dataloader, or hyperparameters are expected
-
Aggregator-based Workflow: Define an experiment and distribute it manually. All participants can verify model code and FL plan prior to execution. The federation is terminated when the experiment is finished
The quickest way to test OpenFL is to follow our tutorials.
Read the blog post explaining steps to train a model with OpenFL.
Check out the online documentation to launch your first federation.
- Ubuntu Linux 16.04 or 18.04.
- Python 3.6+ (recommended to use with Virtualenv).
OpenFL supports training with TensorFlow 2+ or PyTorch 1.3+ which should be installed separately. User can extend the list of supported Deep Learning frameworks if needed.
Federated learning is a distributed machine learning approach that enables collaboration on machine learning projects without having to share sensitive data, such as, patient records, financial data, or classified information. The minimum data movement needed across the federation is solely the model parameters and their updates.
OpenFL builds on the OpenFederatedLearning framework, which was a collaboration between Intel and the University of Pennsylvania (UPenn) to develop the Federated Tumor Segmentation (FeTS, www.fets.ai) platform (grant award number: U01-CA242871).
The grant for FeTS was awarded to the Center for Biomedical Image Computing and Analytics (CBICA) at UPenn (PI: S. Bakas) from the Informatics Technology for Cancer Research (ITCR) program of the National Cancer Institute (NCI) of the National Institutes of Health (NIH).
FeTS is a real-world medical federated learning platform with international collaborators. The original OpenFederatedLearning project and OpenFL are designed to serve as the backend for the FeTS platform, and OpenFL developers and researchers continue to work very closely with UPenn on the FeTS project. An example is the FeTS-AI/Front-End, which integrates UPenn’s medical AI expertise with Intel’s framework to create a federated learning solution for medical imaging.
Although initially developed for use in medical imaging, OpenFL designed to be agnostic to the use-case, the industry, and the machine learning framework.
You can find more details in the following articles:
| Algorithm Name | Paper | PyTorch implementation | TensorFlow implementation | Other frameworks compatibility | How to use |
|---|---|---|---|---|---|
| FedAvg | McMahan et al., 2017 | ✅ | ✅ | ✅ | docs |
| FedProx | Li et al., 2020 | ✅ | ✅ | ❌ | docs |
| FedOpt | Reddi et al., 2020 | ✅ | ✅ | ✅ | docs |
| FedCurv | Shoham et al., 2019 | ✅ | ❌ | ❌ | docs |
We welcome questions, issue reports, and suggestions:
This project is licensed under Apache License Version 2.0. By contributing to the project, you agree to the license and copyright terms therein and release your contribution under these terms.
@misc{reina2021openfl,
title={OpenFL: An open-source framework for Federated Learning},
author={G Anthony Reina and Alexey Gruzdev and Patrick Foley and Olga Perepelkina and Mansi Sharma and Igor Davidyuk and Ilya Trushkin and Maksim Radionov and Aleksandr Mokrov and Dmitry Agapov and Jason Martin and Brandon Edwards and Micah J. Sheller and Sarthak Pati and Prakash Narayana Moorthy and Shih-han Wang and Prashant Shah and Spyridon Bakas},
year={2021},
eprint={2105.06413},
archivePrefix={arXiv},
primaryClass={cs.LG}
}