The models in this repo are based on: From Excitatory and Inhibitory Interactions in Localized Populations of Model Neurons Hugh R. Wilson and Jack D. Cowan; Biophys J. 1972 January; 12:1-24. doi:10.1016/S0006-3495(72)86068-5
The Wilson and Cowan model describes the dynamics and interaction between the excitatory and inhibitory population of neurons.
Following the Bard Ermentrout's implementation in xppau, the dynamics of the model is defined as:
where u and v represents the proportion of excitatory and inhibitory cells firing, i_e and i_i are input currents, z_e and z_i correspond to constant modulatory currents that are applied to the populations, wee, wei, wie, wii are the connections synaptic coupling strength between the excitatory-excitatory, excitatory-inhibitory, inhibitory-excitatory, and inhibitory-inhibitory population respectively.
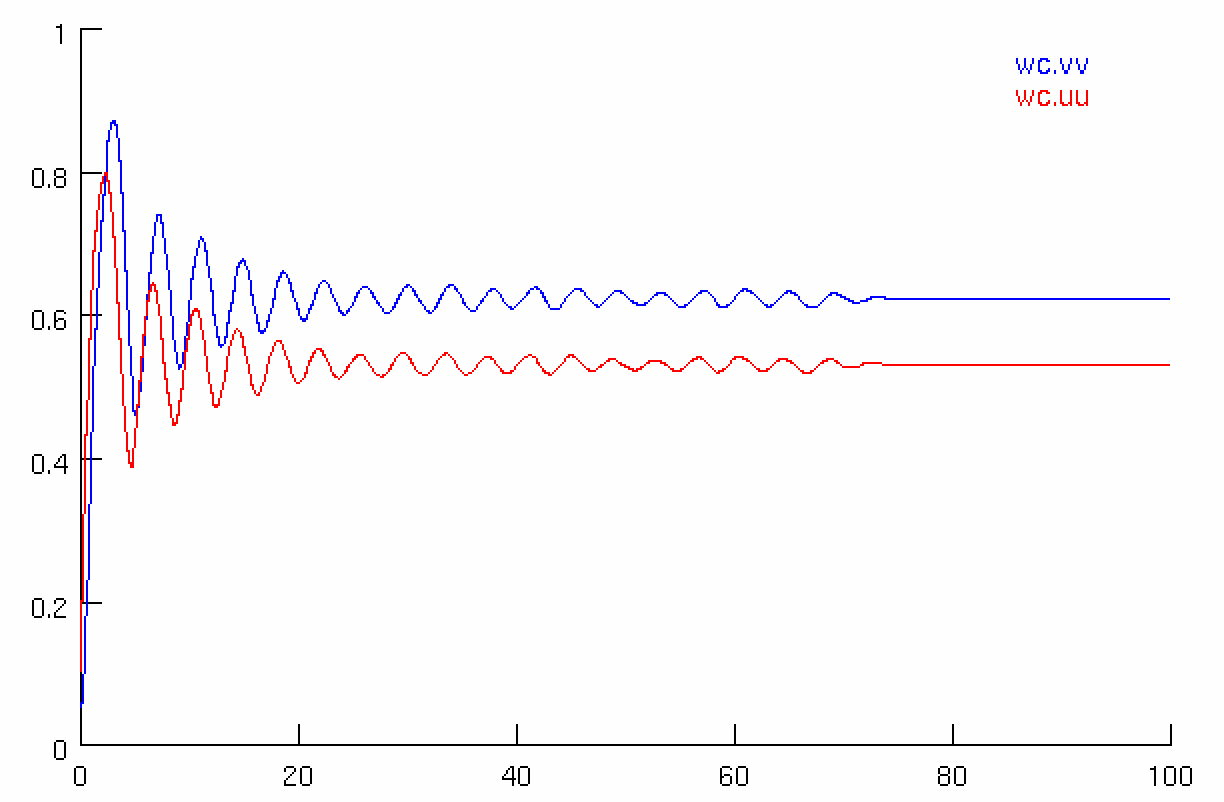
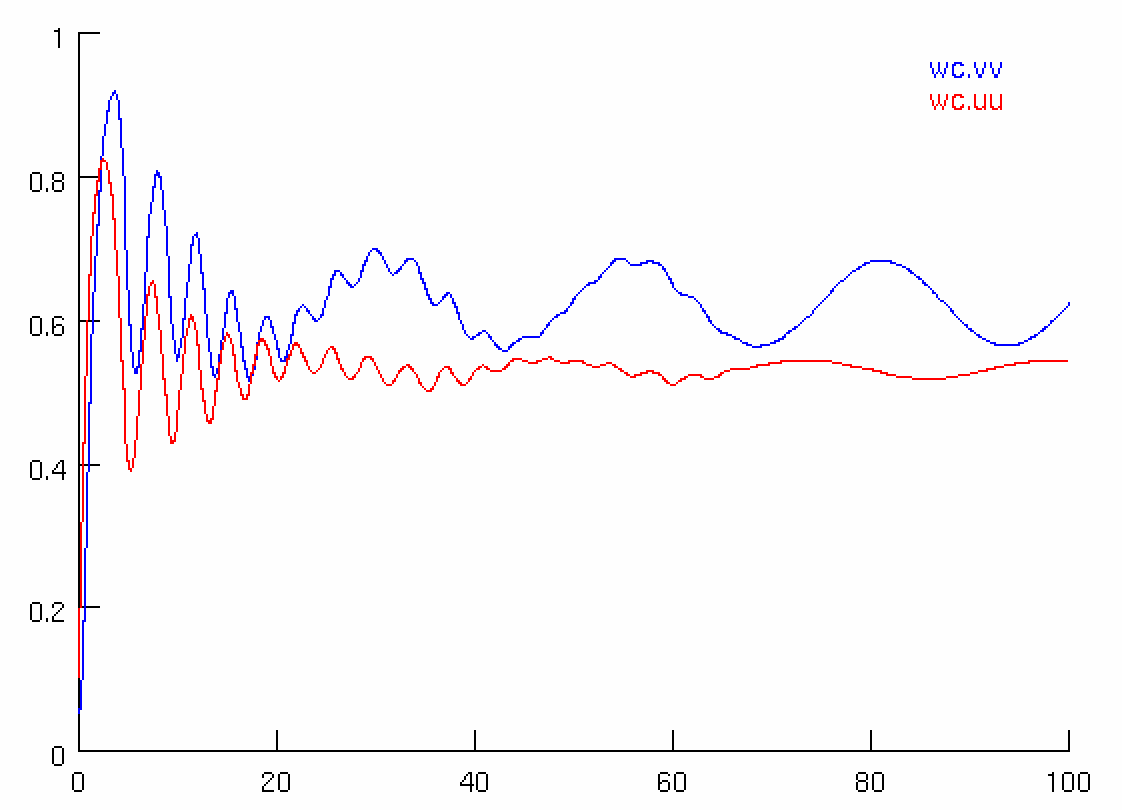
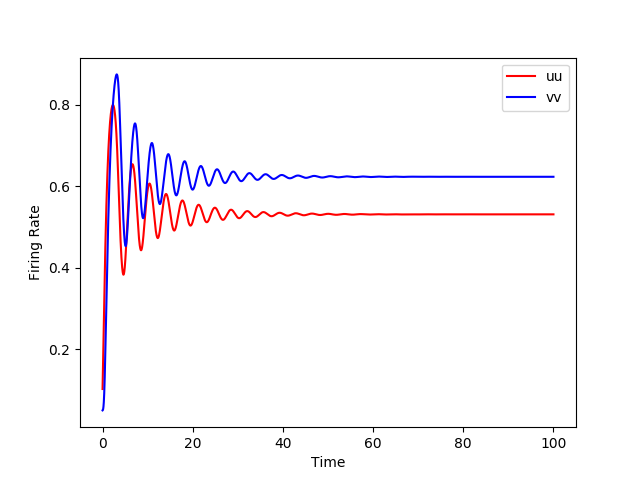
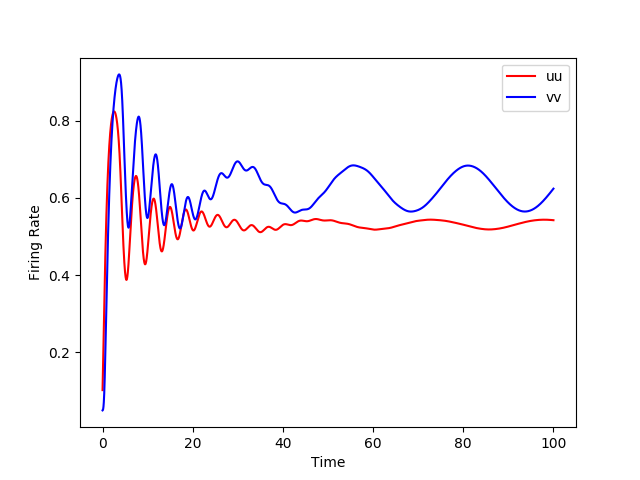
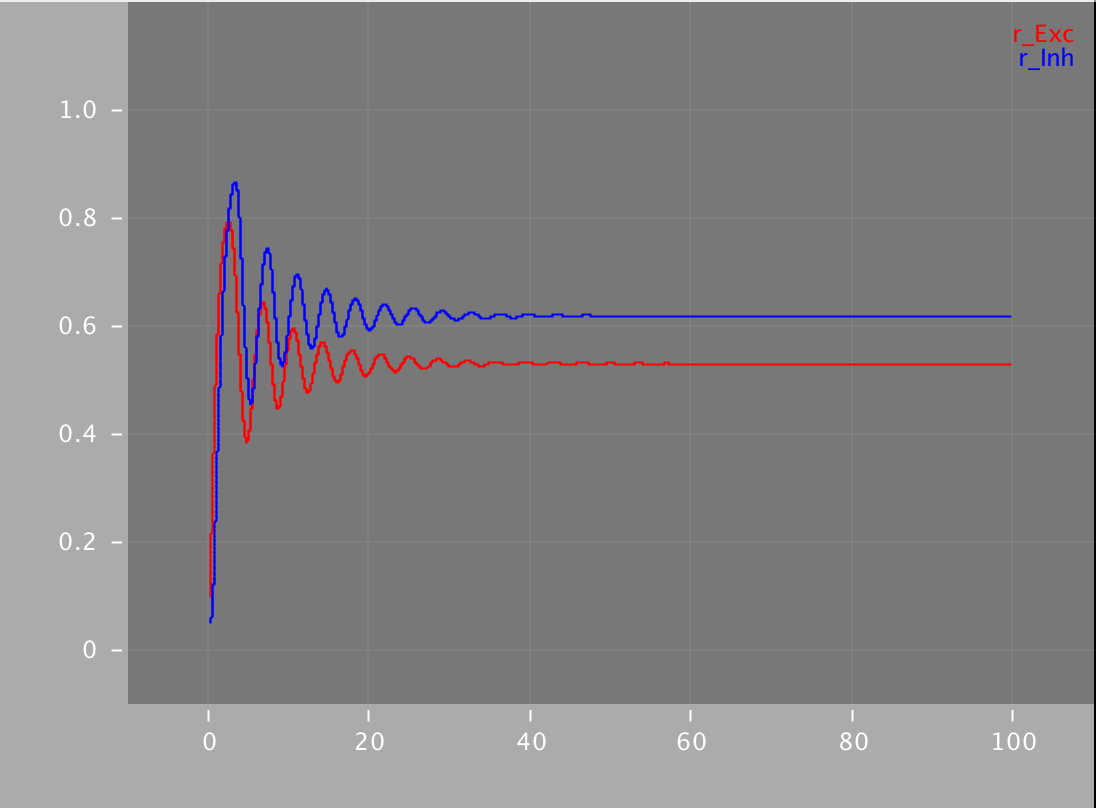
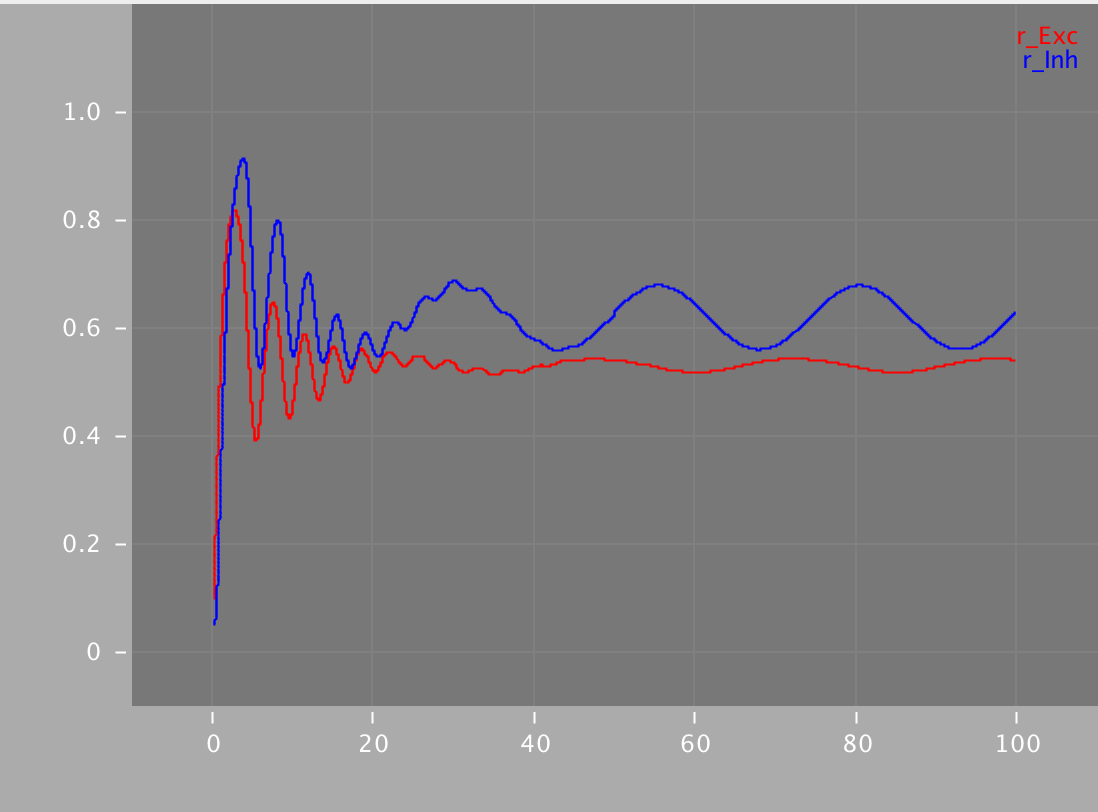
Simulations for the Wilson and Cowan model are available in XPP, NEURON, Python and NeuroML. Using the different simulators we analysed the dynamics with (right images below) and without (left images below) an additional sinusoidal input current.
Although we did not analyse this simulation, for completeness the links for the XPP can be found here.
See here.
To run the simulation on the NEURON folder type:
nrnivmodl
nrngui wc.hoc
To run the simulation on the Python folder type:
python WilsonCowan.py -wee 10. -wei 12. -wie 8. -wii 3. -ze 0.2 -zi 4. -ie1 0 -ii1 0 -w 0.25
python WilsonCowan.py -wee 10. -wei 12. -wie 8. -wii 3. -ze 0.2 -zi 4. -ie1 0.5 -ii1 0.5 -w 0.25
To run the simulation on the NeuroML2 folder type:
jnml LEMS_WC_slow.xml
jnml LEMS_WC_driven.xml
We also explored the generated dynamics of the models in NeuroML2 and Python.
While a more detailed description of the dynamics and isoline analysis with different parameters can be found here, a short example of isocline analysis in NeuroML2 can be found in this [jupyter notebook].